Darren Monckton from the University of Glasgow spoke about the phenomenon of somatic instability (SI) in the context of disease progression. He described the impact of rare polymorphisms (present in 1.4% of individuals) in HTT CAG tract where some CAA sequence variants can affect the age of onset, using a large 3500 samples from the Enroll-HD cohorts. The message is that what seems significant is the number of contiguous CAG repeats in the DNA, not the number of glutamine amino acids at the protein level.
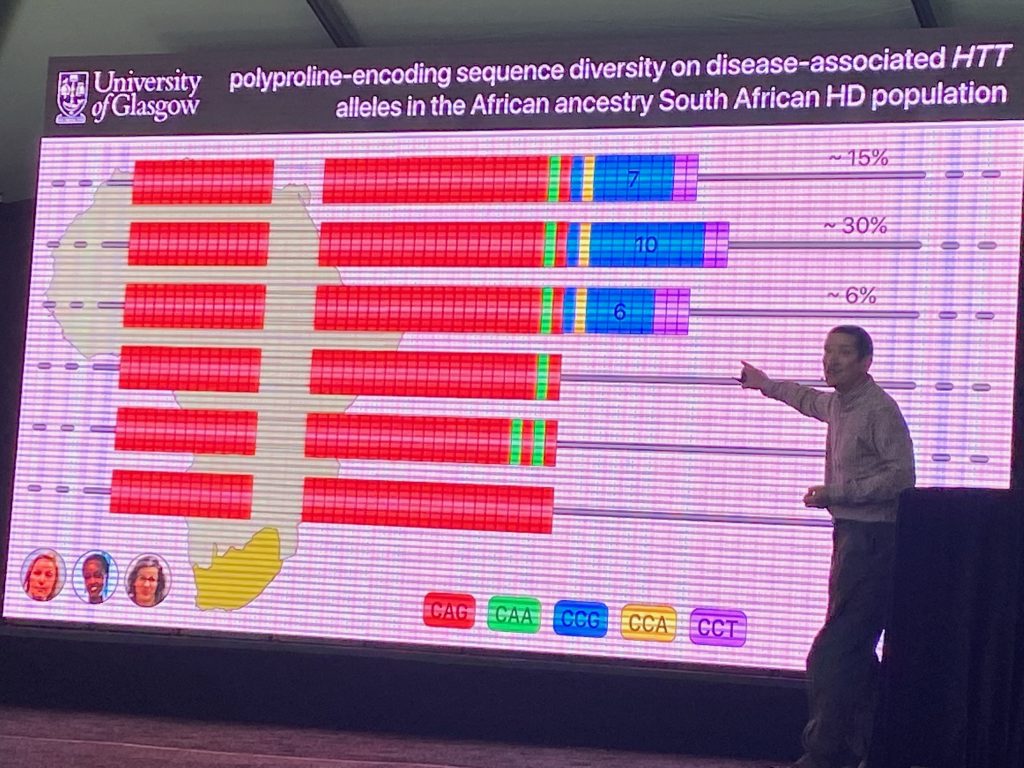
However, more recent work has pointed to other rare changes in the HTT exon-1 region that affects the initial rate of disease progression (the age of onset of disease). Darren’s team sequenced HTT in HD individuals from South Africa, where genetic diversity is higher than in European populations. Using this population genomic information, he identified DNA polymorphisms in the polyproline stretch (which can vary between 7-12 CCG residues, for instance), which were shown to impact the age of onset, and which are rarely found in DNA samples from European populations. Some of the polymorphisms (such as the loss of a CCA proline triplet) were shown to impact age of onset by about 10 years. This change was shown also in a large European population to advance disease onset. His work argues that the polyproline polymorphism involving the loss of the CCA codon, but not the length of the CCG repeats, have the most significant effect on the age of onset and that this effect involves a mechanism independent of SI.
Darren spoke about the potential mechanism by which the loss of the CAA might lead to these profound changes, and speculated that the structure of the RNA might be affected by this loss, as well as by the CAA interruptions. This is an area that requires more work.
STEVE McCARROLL
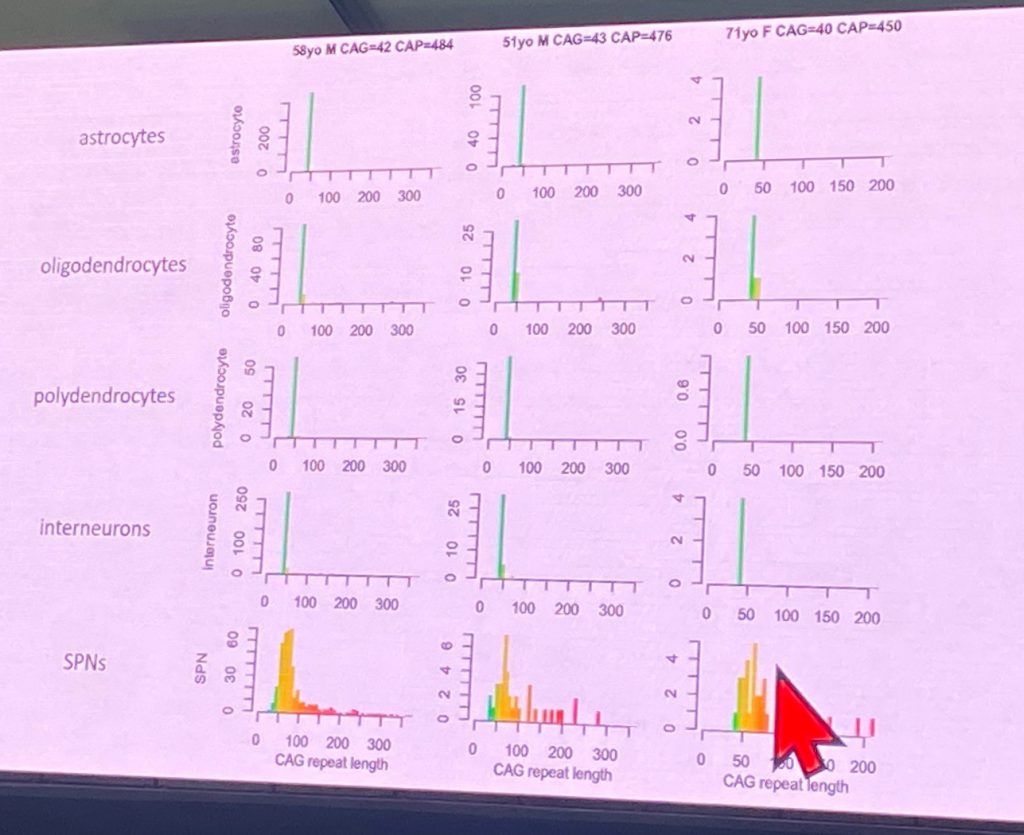
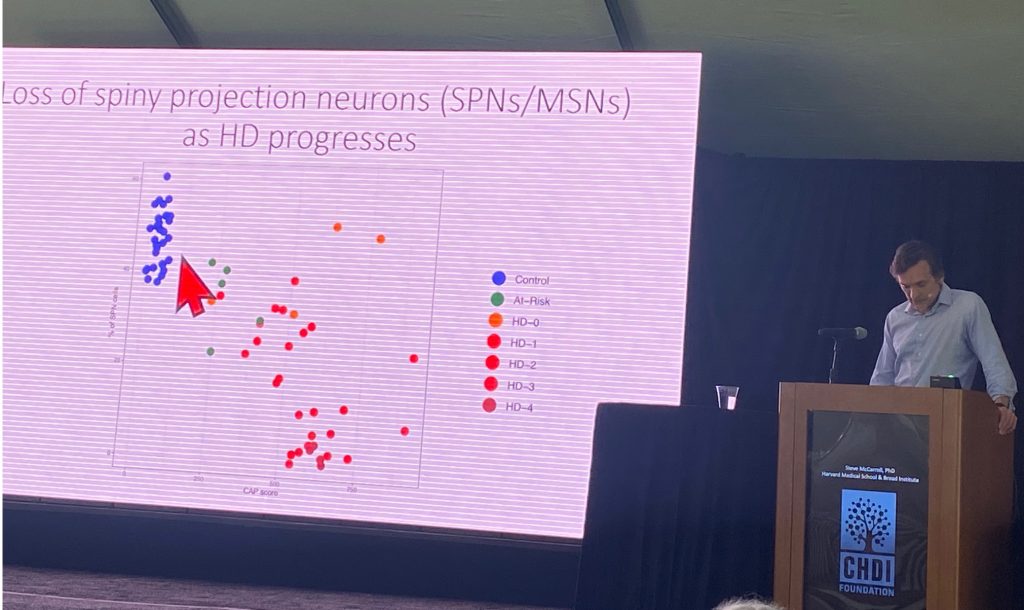
Steve McCarroll from Harvard University, spoke about his work with single cell sequencing of human HD brain cells, in work funded by CHDI Foundation. This novel technique (http://drop-seq.org) allows his lab to isolate and monitor transcriptional changes in a multitude of brain cells, most of which have never been studies in HD brains. Right now, he has collected data from 41 control brains and 42 HD brains ( n=5 from at-risk individuals, n=4 from stage HD-0, n=11 from stage HD-1, n=8 from HD-2, n=9 from HD-3 and n=5 from stage HD-4). Steve described the profound loss of spiny projection neurons (SPNs) with CAP scores (eg as disease progresses), as shown below
Based on his work, Steve shows that HD progression involves changes in every cell type so far evaluated, including in glia and endothelial (blood vessel) cells. How to interpret these changes is challenging due to the loss of cells found as disease progresses. Steve also describe the study of expression QTLs (quantitative trait loci) in terms of RNA levels, as well as the genetic modifiers identified for age of onset, such as Msh3, Fan1, etc. For example, the Msh3 eQTL has an effect of about 10% for Msh3 RNA expression in neurons and astrocytes. Steve then turned to measure CAG repeat sizes in single nuclei isolated from HD brains, which allows his lab to understand the extent of somatic expansion at the HTT gene across cell types and disease stages. The somatic expansion (including expansions up to 350 CAG repeats) its found in striatal SPNs and also in cortical glutamatergic neurons, but not in striatal interneurons. However cortical interneurons do show significant expansion. So it seems that context matters more than cell identity for somatic expansion.