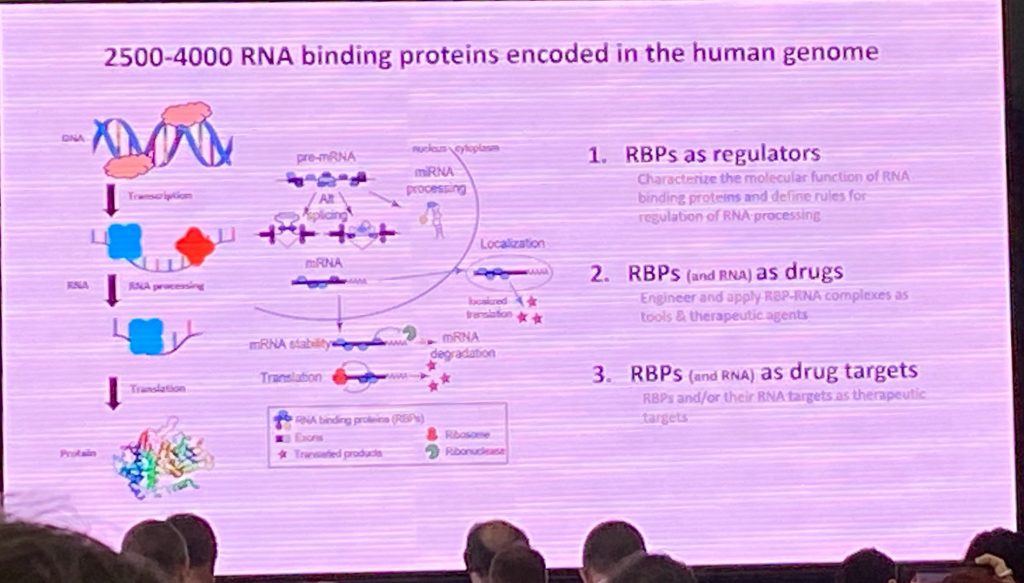
Dr. Gene Yeo from UCSD spoke about his recent work on targeting RNA degradation using Crispr-Cas13d. His lab focused on RNA binding proteins (RBPs) as regulators of gene expression, and as drug targets. He has developed methods to systematically study RBP functions, such as the enhanced eCLIP method, which his lab has done to characterize >200 RBPs. He has also developed methods to track RNA using live imaging in single cells. His methods have been used to target repeat expansion RNAs using Crispr-Cas technology. Recently he has focused on using the Cas13d enzyme to degrade specific RNAs, including HTT by targeting the CAG repeat. The Cas13d/cripsr system can fit into AAVs and therefore can be used for in vivo studies without having to use a dual vector system. He showed that this system can degrade the HTT RNA and correct some of the molecular alterations found in HD iPSCs, in a collaboration between his lab and the lab of Dr. Leslie Thompson.
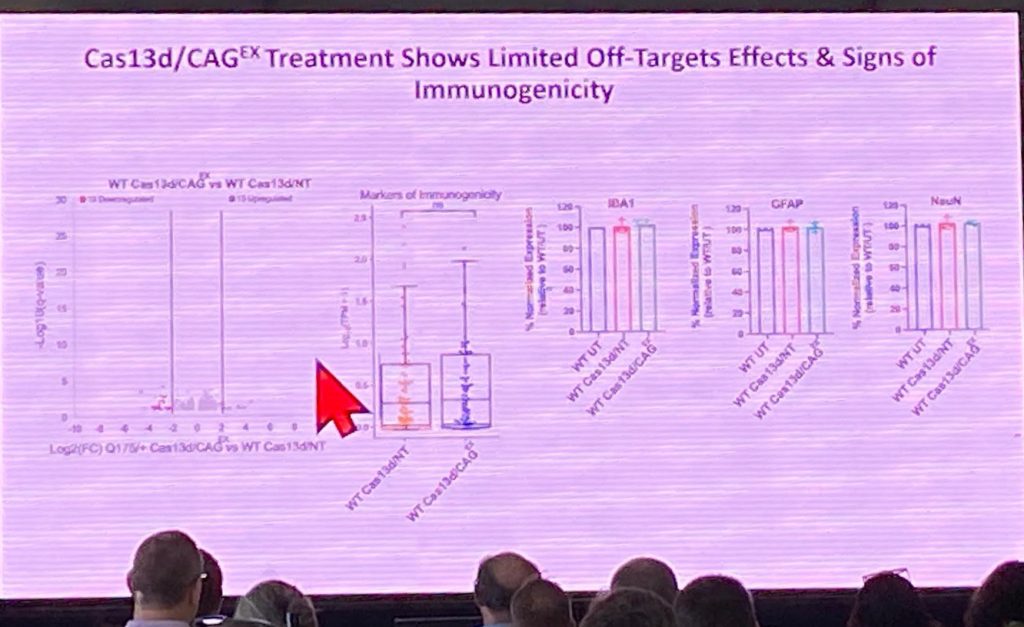
Gene went on to describe that his system has fewer off target effects on other CAG-containing repeat genes. Rodent experiments with AAV-Cas13d/CAGex in Q175s can ameliorate behavioral effects in this model, decrease HTT aggregate pathology, and also restore striatal volume. The effect on the Q175s seemed to be allele specific, as no changes in wildtype mouse HTT were detected by Western blots, with no evidence of inflammation or off-target effects. His lab is now focusing on other human RBPs to improve human therapeutic development. Significant additional analysis is lacking in my opinion in terms of specificity of the approach and its applicability for therapeutic development, but this approach is significant in that it targets the RNA rather than the DNA using crispr (therefore not permanently editing the genome, including unintended genomic locations), enabling potential allele-specific targeting that might be better tolerated.
Dr. Irina Antonijevic from Triplet Therapeutics described their work on developing a therapeutic antisense oligonucleotide (ASO) targeting the gene Msh3, a critical gene that controls somatic expansion of the HTT CAG repeat in the brain, as identified by GWA (genome-wide association) studies. Initially, Msh3 was shown to regulate somatic instability of the HTT gene in mice, and the human genetic association studies supported the conservation of Msh3 function in controlling the rate of expansion of the CAG repeat and the age of onset of motor diagnosis in HD individuals.
The SHIELD-HD study is a natural history study conducted by Triplet which has enrolled 70 individuals in 9 sites in 5 countries (USA, Canada, France, Germany and UK). The study will have a duration of 2 years and will support the planned Phase 1/2a study with the ASO. Irina described the initial data on the structural MRI changes in caudate volume and ventricular volume over the longitudinal study over a period of 48 weeks, suggesting that imaging might be suitable as a biomarker endpoint to track. The PIN HD measurements includes a composite of the total motor score, SDMT (single digit modality test), and rater scores, and also showed changes over the period being monitored so far. They are also measuring CSF levels of NFL (neurofilament light chain), but she did not report results.

Irina then described recent work in NHPs the work done to advance TTX-3360, the clinical candidate ASO. The extent of Msh3 suppression after ICV administration is large and sustained over a 24-week period (>50% suppression is the target for Msh3, based on the work in rodents suggesting this level of suppression should modulate SI of the HTT CAG repeat). Finally, they described the biomarker work done on TTX-3360 using CSF measurements of Msh3 mRNA contained inside brain-derived exosomes (extracellular vesicles). The SHIELD-HD samples of CSF were essential for assay development to detect Msh3 levels and changes after ASO administration. The final platform included qPCR analysis for mRNA detection in human and primate CSF. Housekeeping control genes include HKG, ActB and EEF2 for normalization of the changes in the CSF after treatments. The use of these control genes were corroborated with the CSF samples from the SHIELD-HD study, ensuring no change in untreated individuals over time. In the NHP studies, cisterna magna collection of CSF was better for Msh3 detection over collections via intrathecal (lumbar) collection of CSF. This is pending confirmation in human collection studies. They also measured healthy and HD Msh3 expression in CSF vesicles, which did not change over disease (but the sample size was small). There is a 2/3-fold variability in expression across the individuals measured. No longitudinal data has been analyzed yet, although this is a critical area of work.
Incidentally, they report that the size distribution of the extracellular vesicles isolated from CSF change in the course of disease – so this is an aspect they want to pursue further. No data was presented yet in vivo, that dosing of the ASO changes Msh3 levels in CSF EVs that correlate with parenchymal lowering of the target. This is the critical study that is pending.




